Rationale and Protocol
The pBullet vector series addresses some of the issues with existing localization vectors, namely:
- Size: ~8000 bp including ccdb region (Gateway®), after recombination ~6000 bp plus gene of interest
- Yield: The vector backbone was constructed with the high copy origin of replication from pUC and results in tens of ug from an overnight 5mL culture.
- Modular Design: A series of unique restriction sites were incorporated as well as a PmII site between two resistance markers (AmpR / KanR) to allow another marker to be inserted. Most components can be excised from the vector using restriction enzymes.
- High Throughput: The pBullet vectors employ the Gateway® recombination technology that enable fast creation of constructs.
- Co-localization: fluorescent marker in the plasmid enabling simultaneous localization of gene of interest and organelle verification.
- Markers: complete coverage of the endomembrane cytosol, ER, cis-Golgi, trans-Golgi, endosome, vacuole and plasma membrane
- N or C-terminal: localization of gene of interest can be conducted using a C-terminal or N-terminal EYFP providing higher confidence for subcellular location
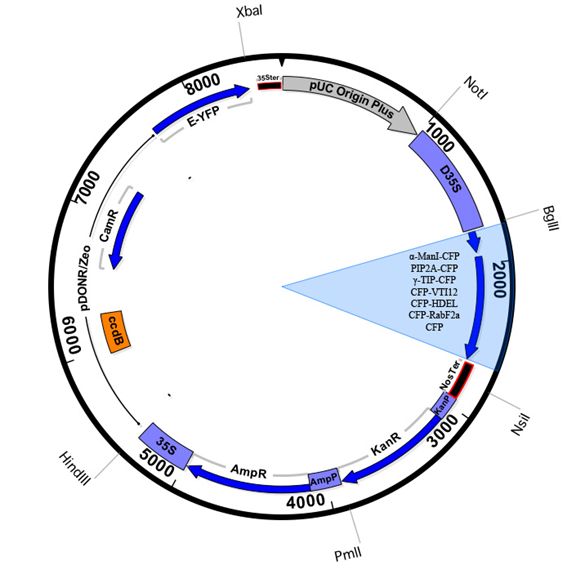
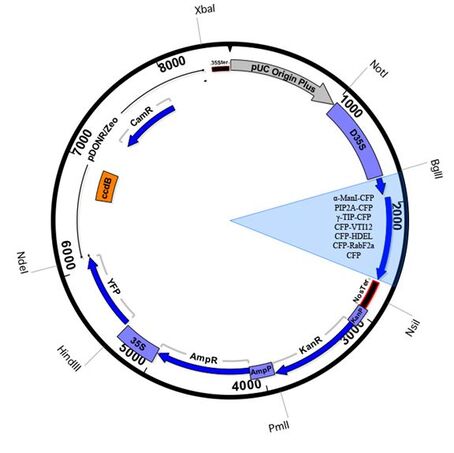
Vector Maps
Plasmid Name |
Marker (ECFP) |
Location |
Gene |
Vector Maps (n) |
Vector Maps (c) |
Reference |
pBullet-cyt-n/c |
ECFP |
cytosol |
n/a |
|||
pBullet-er-n/c |
WAK2 (29 aa)-ECFP-HDEL |
ER |
||||
pBullet-cg-n/c |
alpha-Man I (49 aa)-ECFP |
cis-Golgi |
||||
pBullet-tgn-n/c |
ECFP-VTI12 |
trans-Golgi |
||||
pBullet-end-n/c |
ECFP-RabF2a |
endosome |
||||
pBullet-vac-n/c |
gamma-TIP-ECFP |
vacuole |
||||
pBullet-pm-n/c |
PIP2a-ECFP |
plasma membrane |
Biolistics Protocol
The approach employed for the biolistic transformation of onion essentially follows the protocol outlined by Rech et al., (2008) with some minor modifications.
Equipment
Microcarrier preparation
Coating microcarriers with DNA
Bombardment
Equipment
- PDS-1000/He™ System (Bio-Rad, USA)
- Fresh medium-sized brown onions
- Gold particles, microcarrier and other biolistic consumables (Bio-Rad, USA)
Microcarrier preparation
- Weigh out 15 mg of gold particle (1µm diameter, Bio-Rad, USA)
- Add 1mL 80% EtOH
- Vortex 3 min
- Let microcarrier soak for 15 min
- Quick spin for 5 sec
- Discard supernatant
- Wash with sterile water 3 times.
- Resuspend in 500 µL sterile 50% glycerol
- Aliquot 25 µL per tube
Coating microcarriers with DNA
- Add 2.5µL of DNA (at least 200ng/µL)
- Add 25µL of 2.5M CaCl2
- Add 10µL of 0.1M spermidine
- Vortex for 10 min at 3000 RPM
- Quick spin for 5 sec
- Discard Supernatant
- Wash supernatant with 140µL of 100% ethanol
- Resuspend with 20µL of 100% ethanol
Bombardment
- Wash macrocarrier and pressure disk (1100 PSI) with 100% ethanol
- Load 20µL microcarrier onto macrocarrier
- Allow to dry (takes about 15-30 min)
- Set up PDS-1000/He system according to manufacturer’s recommendations (vacuum of 28 inch/Hg, target distance of 6cm, and helium pressure of 1100psi). We used the original rupture disk retaining cap with the hepta adapter macrocarrier holder. The macrocarrier is placed on the center holder, leaving the other six empty.
- Keep onion in a moist plate, in the dark, and leave overnight before visualizing
|
Standard macrocarrier holder (left) and hepta adapter macrocarrier holder (right). We use a hybrid setup that employs the standard rupture disk retaining cap with the central ring on the hepta adapter macrocarrier holder.
|
Bio-Rad biolitics apparatus with the hepta adapter loaded with onion epidermal samples loaded at the 6cm point.
|
Examples
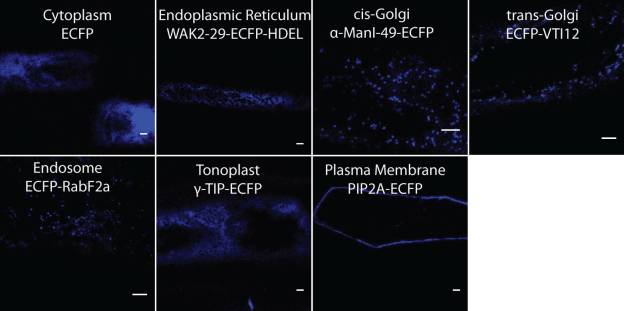
The current set of validated organelle markers (ECFP) in the pBullet collection.
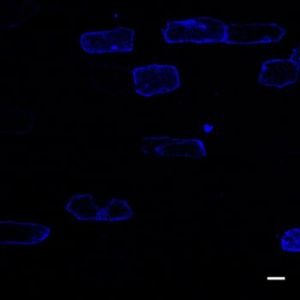
Transformation efficiency of a typical bombardment with 1 ug pBullet plasmid. Scale = 10 um.